Transposcope is a package that is used to evaluate potential retrotransposon insertions, which have been identified by either MELT or TIPseqHunter (A generic input type will be added in later releases).
As novel insertions are not present within the reference genome, reads are unable to be aligned correctly. TranspoScope solves this problem by creating a localized reference sequence that matches what was proposed by either MELT or TIPseqHunter. Reads which aligned to regions flanking the novel insertion are then realigned to the localized sequence.
TranspoScope includes functionality to visualize and assess the realigned regions within a web-based visualization.
- If Anaconda is not currently installed, install Miniconda using the installer and instructions available here, https://docs.conda.io/en/latest/miniconda.html#miniconda.
- Set up the Bioconda channels as described in the following link, https://bioconda.github.io/#set-up-channels.
Run the following command in the terminal:
conda create -n transposcope_env python=3.7 transposcopeThis command will create a new conda environment called 'transposcope_env' which uses Python 3.7 and includes the transposcope package.
Alternatively, TranspoScope can be installed into the current conda environment using the following command.
conda install transposcope=2.0.1https://github.com/FenyoLab/transposcope/wiki
TranspoScope has been tested on Linux and OSX. Realignment is not possible using regular conda on Windows as the Samtools conda package is not available on this platform. Viewing files which are hosted online will work on any operating system as long as a modern web browser is used.
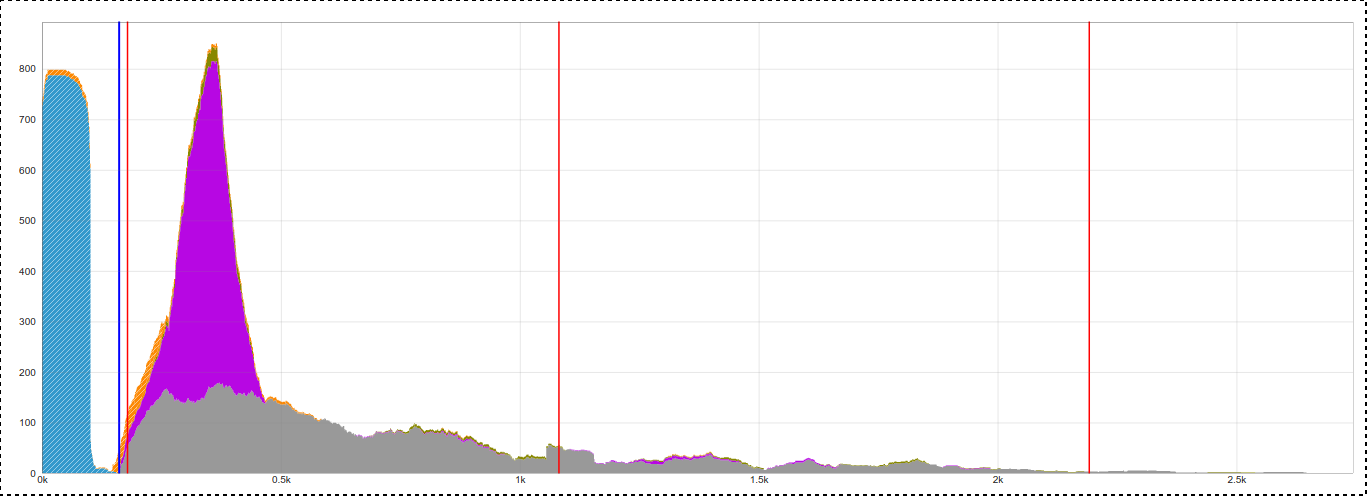
This example shows strong evidence as there are ~800 bridging reads where one read aligns with the reference sequence, and the pair aligns with the LINE-1 reference.
Interactive visualization link
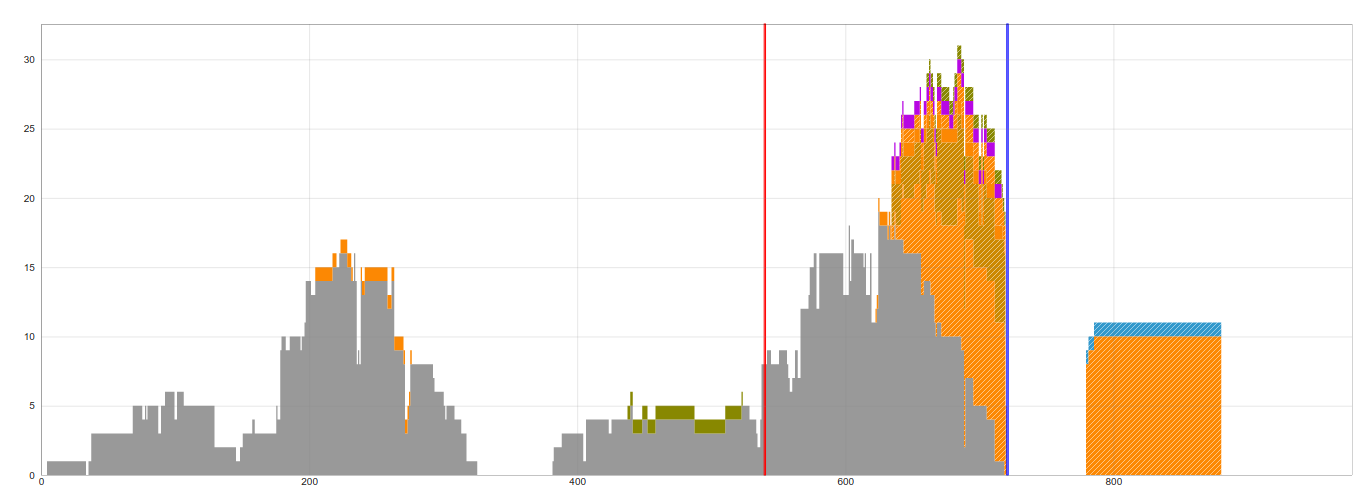
This example is more difficult to classify as there are only one bridging reads and 10 junction reads supporting the insertion.
Interactive visualization link
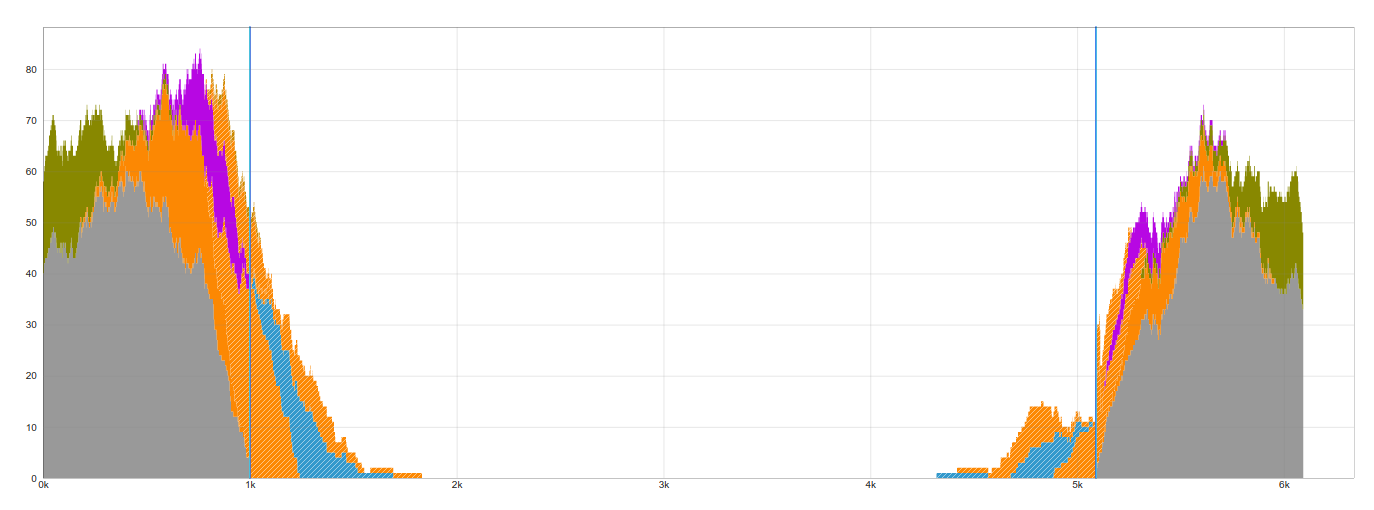
This example shows strong evidence of there being an insertion. There are many bridging reads on both the 3' and 5' ends of the insertion, as well as many junction reads, which match the proposed LINE-1 reference sequence.
Interactive visualization link
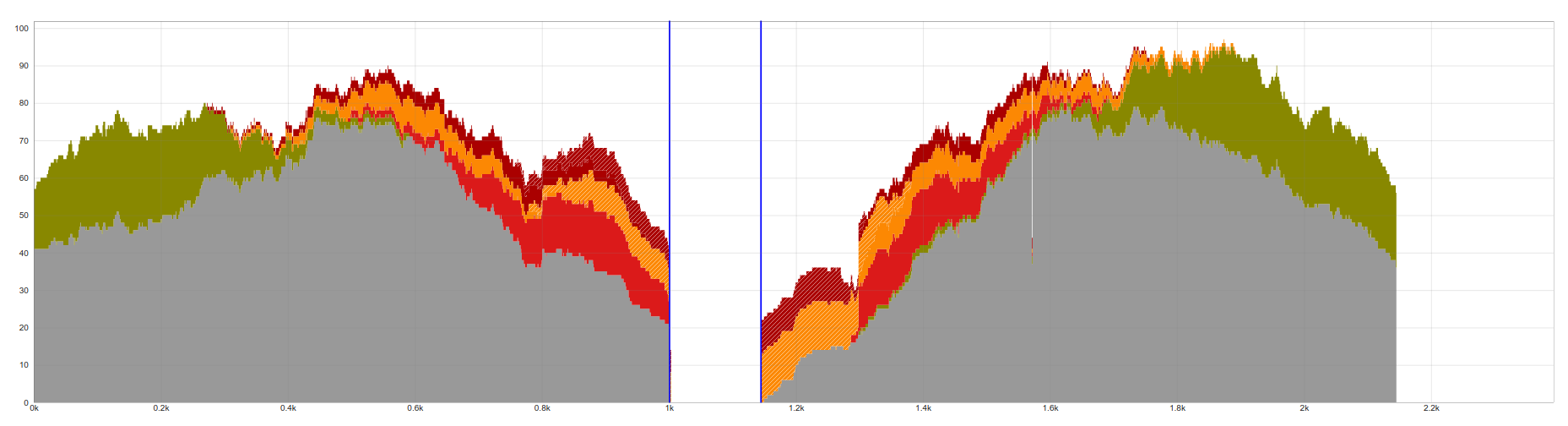
This example shows little evidence that there really is an insertion at this locus. Junction reads on the 5' flanking side match 3 bases of the proposed LINE-1 reference sequence with no other matches.