-
Notifications
You must be signed in to change notification settings - Fork 46
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
TriangularMesh #569
Comments
|
Current work at https://github.com/magpylib/magpylib/tree/trimesh_body Tasks
Other tasks
|
|
@Alexboiboi I played a bit around with Points of Discussion
what do you think ? |
|
Discussion @OrtnerMichael and @Alexboiboi :
Checks
Graphics
|
|
Hi @OrtnerMichael, The changes I could implement are now up to date with test (100% coverage). Waiting for feedback. The styling topic is mostly independent, and I'll continue to work on in in another branch |
|
Looks great - everything works as discussed ! import pyvista as pv
import magpylib as magpy
magpy.magnet.TriangularMesh.from_pyvista(
magnetization=(1,2,3),
polydata=pv.Text3D("I'm loving it")
).show(backend='pyvista')I'll check and work on the methods face_flip, inside_outside, closed_mesh, separate_bodies, and also to clean up e.g. checks that are in the TriangularMesh class ... not sure if they should be there... and maybe start working on the docstrings. |
|
Hi @OrtnerMichael , I have updated the style structure to allow for composition. The Can you check it out? import magpylib as magpy
import pyvista as pv
trimesh = magpy.magnet.TriangularMesh.from_pyvista(magnetization=(0, 0, 1000),polydata=pv.Cube())
trimesh._triangles = trimesh.triangles[1:] # open the mesh
trimesh.show(backend="plotly")import magpylib as magpy
import pyvista as pv
import numpy as np
mag = (111, 222, 33)
disjoint_mesh = magpy.magnet.TriangularMesh.from_pyvista(mag, pv.Text3D("ABC"))
disjoint_mesh.show(backend="plotly") |
|
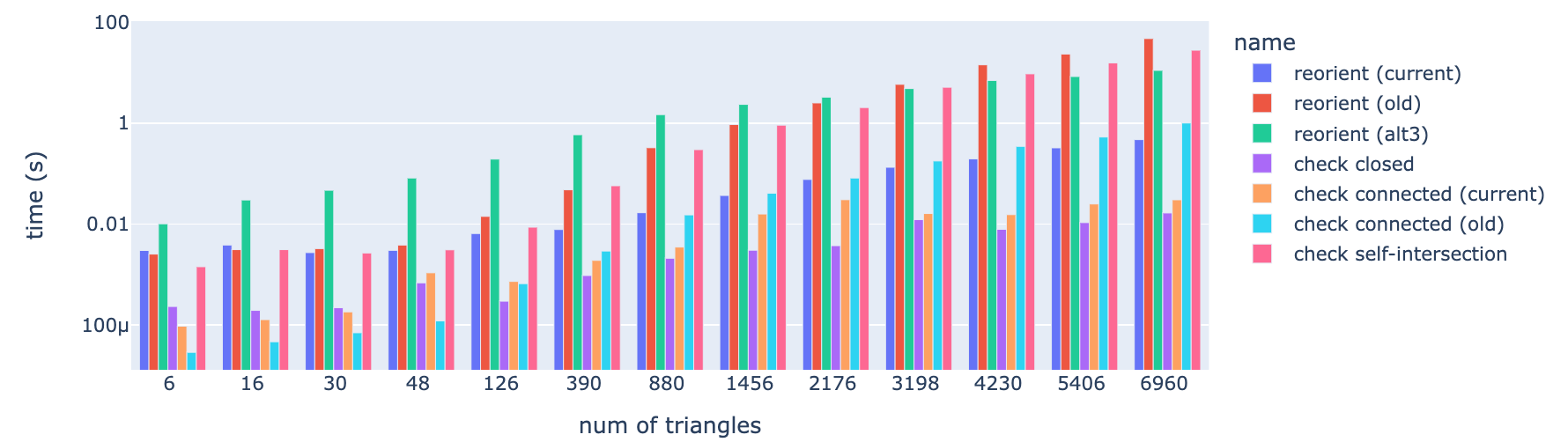
Hi @OrtnerMichael, I have done some basic profiling of the implemented mesh validation algorithms (also including self-intersection). To reproducefrom time import perf_counter
import magpylib as magpy
import numpy as np
import pandas as pd
import plotly.express as px
import pyvista as pv
from magpylib._src.fields.field_BH_triangularmesh import (
fix_trimesh_orientation,
get_disjoint_triangles_subsets,
lines_end_in_trimesh,
mask_inside_trimesh,
trimesh_is_closed,
v_norm_cross,
)
def trimesh_is_connected_old(triangles: np.ndarray) -> np.ndarray:
"""
Check if triangular mesh consists of multiple disconnected parts.
Input: triangles: np.ndarray, shape (n,3), dtype int
triples of indices
Output: bool (True if connected, False if disconnected)
"""
tri_list = triangles.tolist()
connected = [np.min(triangles)]
# start with lowest index
# cycle through tri_list and unique-add indices from connecting triangles
# when adding indices to connected, remove respective triangle from tri_list
# After completing a cycle, check if a new triangle was added
# if not, and tri_list still contains some triangles, then these faces must be
# disconnected from the rest
added_new = True
while added_new:
added_new = False
# cycle through all triangles that are not connected yet
for tria in tri_list:
for ii in tria:
if ii in connected:
# add new triangle to connected
for iii in tria:
if iii not in connected:
connected.append(iii)
tri_list.remove(tria)
added_new = True
break
if tri_list:
return False
return True
def fix_trimesh_orientation_old(vertices: np.ndarray, triangles: np.ndarray) -> np.ndarray:
"""
Check if all triangles are oriented outwards. Fix the ones that are not, and return an
array of properly oriented triangles.
Parameters
----------
vertices: np.ndarray, shape (n,3)
vertices of the mesh
triangles: np.ndarray, shape (n,3), dtype int
triples of indices
Returns
-------
triangles: np.ndarray, shape (n,3), dtype int
fixed triangles
"""
facets = vertices[triangles]
# compute facet orientations (normalized)
a = facets[:, 0, :] - facets[:, 1, :]
b = facets[:, 1, :] - facets[:, 2, :]
orient = v_norm_cross(a, b)
# create check points by displacing the facet center in facet orientation direction
eps = 1e-6 # unfortunately this must be quite a large number :(
facet_center = facets.mean(axis=1)
check_points = facet_center + orient * eps
# find points which are now inside
inside_mask = mask_inside_trimesh(check_points, facets)
# flip triangles which point inside
triangles[inside_mask] = triangles[inside_mask][:, [0, 2, 1]]
return triangles
def fix_trimesh_orientation2(vertices: np.ndarray, triangles: np.ndarray) -> np.ndarray:
"""
Check if all triangles are oriented outwards. Fix the ones that are not, and return an
array of properly oriented triangles.
Parameters
----------
vertices: np.ndarray, shape (n,3)
vertices of the mesh
triangles: np.ndarray, shape (n,3), dtype int
triples of indices
Returns
-------
triangles: np.ndarray, shape (n,3), dtype int
fixed triangles
"""
facets = vertices[triangles]
# compute facet orientations (normalized)
a = facets[:, 0, :] - facets[:, 1, :]
b = facets[:, 1, :] - facets[:, 2, :]
orient = v_norm_cross(a, b)
# create check points by displacing the facet center in facet orientation direction
eps = 1e-2 # unfortunately this must be quite a large number :(
facet_center = facets.mean(axis=1)
check_points = facet_center + orient * eps
check_points += b * eps # avoid test_line to cross edge
# create test-lines from outside to test-points
facets_mins = np.min(facets, axis=1)
facets_maxes = np.max(facets, axis=1)
for ind, _ in enumerate(triangles):
facet_min, facet_max = facets_mins[ind], facets_maxes[ind]
coords = np.array([0, 1, 2])
first_non_zero_coord = coords[orient[ind] != 0][0]
coords = np.delete(coords, first_non_zero_coord)
pt = check_points[ind]
line = np.array([pt, pt])
line[0, first_non_zero_coord] = facet_min[first_non_zero_coord] - 1
mask_min1 = (facet_min[coords[0]] <= facets_mins[:, coords[0]]) & (
facets_mins[:, coords[0]] <= facet_max[coords[0]]
)
mask_max1 = (facet_min[coords[0]] <= facets_maxes[:, coords[0]]) & (
facets_maxes[:, coords[0]] <= facet_max[coords[0]]
)
mask_min2 = (facet_min[coords[1]] <= facets_mins[:, coords[1]]) & (
facets_mins[:, coords[1]] <= facet_max[coords[1]]
)
mask_max2 = (facet_min[coords[1]] <= facets_maxes[:, coords[1]]) & (
facets_maxes[:, coords[1]] <= facet_max[coords[1]]
)
mask = (mask_min1 | mask_max1) & (mask_min2 | mask_max2)
if lines_end_in_trimesh(np.array([line]), facets[mask])[0]:
triangles[ind] = triangles[ind][[0, 2, 1]]
return triangles
def signed_volume(a, b, c, d):
"""Computes the signed volume of a series of tetrahedrons defined by the vertices in
a, b c and d. The ouput is an SxT array which gives the signed volume of the tetrahedron
defined by the line segment 's' and two vertices of the triangle 't'."""
return np.sum((a - d) * np.cross(b - d, c - d), axis=2)
def segments_intersect_triangle(segments, facets, summation=True):
"""For each line segment in `segments`, this function computes how many times it intersects
any of the facets given in `facets`.
Parameters
----------
segments: Sx2x3 array
Array of `S` line segments where the first index specifies the S^th line segment, the
second index refers to the start or end point of the segment, and the third index points
to the x, y, z coordinates of the line segment point.
facets: Tx3x3
Array of `T` triangular facets, where the first index specifies the T^th triangle, the
second index refers to one of the three vertices (which don't have to be in any particular
order), and the third index points to the x,y,z coordinates the the triangle vertex.
summation: bool, optional
If `True`, a binary array is return which only tells if the S^th line segment intersects
any of the facets given. If `False`, returns a SxT array that tells which facets are
intersected.
Returns
-------
If `'summation'` is `True`, returns a binary array of size S which tells whether the S^th
line segment intersects any of the facets given. Otherwise returns a SxT array that tells
which facets are intersected.
"""
s, t = segments, facets
# compute the normals to each triangle
normals = np.cross(t[:, 2] - t[:, 0], t[:, 2] - t[:, 1])
normals /= np.linalg.norm(normals, axis=1, keepdims=True)
# get sign of each segment endpoint, if the sign changes then we know this segment crosses the
# plane which contains a triangle. If the value is zero the endpoint of the segment lies on the
# plane.
s0, s1 = s[:, :1], s[:, 1:] # -> S x T x 3 arrays
sign1 = np.sign(np.sum(normals * (s0 - t[:, 2]), axis=2)) # S x T
sign2 = np.sign(np.sum(normals * (s1 - t[:, 2]), axis=2)) # S x T
# determine segments which cross the plane of a triangle.
# -> 1 if the sign of the end points of s is
# different AND one of end points of s is not a vertex of t
cross = (sign1 != sign2) * (sign1 != 0) * (sign2 != 0) # S x T
# get signed volumes
v = [
np.sign(signed_volume(t[:, i], t[:, j], s0, s1))
for i, j in zip((0, 1, 2), (1, 2, 0))
] # S x T
same_volume = np.logical_and(
(v[0] == v[1]), (v[1] == v[2])
) # 1 if s and t have same sign in v0, v1 and v2
res = cross * same_volume
if summation:
res = np.sum(res, axis=1)
return res
def check_self_intersection(triangles):
edges = np.concatenate(
[triangles[:, 0:2], triangles[:, 1:3], triangles[:, ::2]], axis=0
)
edges_uniq, edges_counts = np.unique(
np.sort(edges, axis=1), axis=0, return_counts=True
)
facets = vertices[triangles]
segments = vertices[edges_uniq]
return segments_intersect_triangle(segments, facets)
def time_it(func, *args, **kwargs):
start = perf_counter()
func(*args, **kwargs)
return perf_counter() - start
inputs = {
"reorient (current)": {"func": fix_trimesh_orientation, "names": ("vertices", "triangles")},
"reorient (old)": {"func": fix_trimesh_orientation_old, "names": ("vertices", "triangles")},
"reorient (alt3)": {"func": fix_trimesh_orientation2, "names": ("vertices", "triangles")},
"check closed": {"func": trimesh_is_closed, "names": ("triangles",)},
"check connected (current)": {"func": get_disjoint_triangles_subsets, "names": ("triangles", )},
"check connected (old)": {"func": trimesh_is_connected_old, "names": ("triangles", )},
"check self-intersection": {"func": check_self_intersection, "names": ("triangles")},
}
results=[]
tri = []
for resol in [ 3, 4, 5, 6, 9, 15, 22, 28, 34, 41, 47, 53, 60]:
resol = int(resol)
polydata = pv.Sphere(phi_resolution=resol, theta_resolution=resol).triangulate()
vertices = polydata.points
triangles = polydata.faces.reshape(-1, 4)[:, 1:]
print(f"batch with {len(triangles)} triangles")
kwargs = {"triangles": triangles, "vertices": vertices}
for name, val in inputs.items():
t = time_it(val["func"], **{k:v for k,v in kwargs.items() if k in val["names"]})
res = {"name": name, "time": t, "triangles": len(triangles)}
print(res)
results.append(res)
df = pd.DataFrame(results)
df["num of triangles"] = df["triangles"].astype(str) + " "
fig = px.bar(
df,
x="num of triangles",
y="time",
# facet_col="variable",
color="name",
labels={"time": "time (s)"},
#log_x=True,
log_y=True,
facet_col_spacing=0.05,
barmode="group",
)
fig.update_yaxes(showticklabels=True)
fig.show()
fig.update_layout( yaxis_type="linear")
fig.show() |
|
Importing from pyvista leads most of the time to invalid meshes. Especially when performing boolean operations, which is a bumer :( (maybe worth opening a github issue in their repo). It don't see a straightforward solution to this, since we are only performing mesh validation. The only fix performed within magpylib is the reorientation of triangles and we will certainly not start to implement mesh repairing tools within magpylib. We could try with other meshing libraries how boolean operation perform and include some example(s) in the docs if it works well. We could even keep a bad example in the docs for info (see below). @OrtnerMichael any thoughts? import pyvista as pv
import magpylib as magpy
# create a complex pyvista PolyData object
sphere = pv.Sphere(radius=0.85)
dodec = pv.Dodecahedron().triangulate()
obj = dodec.boolean_difference(sphere)
magnet = magpy.magnet.TriangularMesh.from_pyvista(
magnetization=(0,0,100),
polydata=obj,
validate_connected=False,
validate_closed=False,
reorient_triangles=False,
style_label="Dodecahedron cut by Sphere",
)
magnet.show(backend='plotly')import magpylib as magpy
import pyvista as pv
# create a complex pyvista PolyData object
cube = pv.Cube(x_length=0.5, y_length=0.5, z_length=3).triangulate()
cube = cube.rotate_x(30)
dodec = pv.Dodecahedron().triangulate()
obj = dodec.boolean_difference(cube)
magnet = magpy.magnet.TriangularMesh.from_pyvista(
magnetization=(0, 0, 100),
polydata=obj,
validate_connected=False,
validate_closed=False,
reorient_triangles=False,
style_label="Dodecahedron cut by Cuboid",
)
magnet.show(backend="plotly") |
|
Should we add a method |
|
With the help of the import pyvista as pv
import magpylib as magpy
# create a complex pyvista PolyData object
sphere = pv.Sphere(radius=0.85)
dodec = pv.Dodecahedron().triangulate()
obj = dodec.boolean_difference(sphere)
obj = obj.clean()
magnet = magpy.magnet.TriangularMesh.from_pyvista(
magnetization=(0,0,100),
polydata=obj,
validate_connected=False,
validate_closed=False,
reorient_triangles=False,
style_label="Dodecahedron cut by Sphere",
)
magnet.show(backend='plotly')import magpylib as magpy
import pyvista as pv
# create a complex pyvista PolyData object
cyl = pv.Cylinder(radius=0.4, height=2).triangulate()
cyl = cyl.rotate_y(-30)
dodec = pv.Dodecahedron().triangulate()
obj = dodec.boolean_difference(cyl)
obj = obj.clean()
magnet = magpy.magnet.TriangularMesh.from_pyvista(
magnetization=(0, 0, 100),
polydata=obj,
validate_connected=False,
validate_closed=False,
reorient_triangles=False,
style_label="Dodecahedron cut by Cylinder",
)
magnet.show(backend="plotly")import magpylib as magpy
import pyvista as pv
# create a complex pyvista PolyData object
cube = pv.Cube(x_length=0.5, y_length=0.5, z_length=3).triangulate()
cube = cube.rotate_x(30)
dodec = pv.Dodecahedron().triangulate()
obj = dodec.boolean_difference(cube)
obj = obj.clean()
magnet = magpy.magnet.TriangularMesh.from_pyvista(
magnetization=(0, 0, 100),
polydata=obj,
validate_connected=False,
validate_closed=False,
reorient_triangles=False,
style_label="Dodecahedron cut by Cuboid",
)
magnet.show(backend="plotly") |
|
And with the import pyvista as pv
import magpylib as magpy
# create a complex pyvista PolyData object
sphere = pv.Sphere(radius=0.85)
dodec = pv.Dodecahedron().triangulate().subdivide(2)
obj = dodec.boolean_difference(sphere)
obj = obj.clean()
magnet = magpy.magnet.TriangularMesh.from_pyvista(
magnetization=(0,0,100),
polydata=obj,
style_label="Dodecahedron cut by Sphere",
)
magnet.show(backend='plotly')import magpylib as magpy
import pyvista as pv
# create a complex pyvista PolyData object
cyl = pv.Cylinder(radius=0.4, height=2).triangulate()
cyl = cyl.rotate_y(-30)
dodec = pv.Dodecahedron().triangulate().subdivide(2)
obj = dodec.boolean_difference(cyl)
obj = obj.clean()
magnet = magpy.magnet.TriangularMesh.from_pyvista(
magnetization=(0, 0, 100),
polydata=obj,
style_label="Dodecahedron cut by Cylinder",
)
magnet.show(backend="plotly")import magpylib as magpy
import pyvista as pv
# create a complex pyvista PolyData object
cube = pv.Cube(x_length=0.5, y_length=0.5, z_length=3).triangulate()
cube = cube.rotate_x(30)
dodec = pv.Dodecahedron().triangulate().subdivide(2)
obj = dodec.boolean_difference(cube)
obj = obj.clean()
magnet = magpy.magnet.TriangularMesh.from_pyvista(
magnetization=(0, 0, 100),
polydata=obj,
style_label="Dodecahedron cut by Cuboid",
)
magnet.show(backend="plotly") |
|
hi @Alexboiboi
Do you see any other problems when creating complex shapes with pyvista ? Can I review the trimesh body branch ? |
|
U can review the branch, I may just edit som visualization features |
|
To summarize the current validation performance: from time import perf_counter
import magpylib as magpy
import numpy as np
import pandas as pd
import plotly.express as px
import pyvista as pv
from magpylib._src.fields.field_BH_triangularmesh import (
fix_trimesh_orientation,
get_disjoint_triangles_subsets,
trimesh_is_closed,
)
def time_it(func, *args, **kwargs):
start = perf_counter()
func(*args, **kwargs)
return perf_counter() - start
inputs = {
"reorient": {"func": fix_trimesh_orientation, "names": ("vertices", "triangles")},
"validate closed": {"func": trimesh_is_closed, "names": ("triangles",)},
"validate connected": {"func": get_disjoint_triangles_subsets, "names": ("triangles", )},
}
results=[]
tri = []
for resol in [ 3, 4, 5, 6, 9, 15, 22, 28, 34, 41, 47, 53, 60, 67, 75]:
resol = int(resol)
polydata = pv.Sphere(phi_resolution=resol, theta_resolution=resol).triangulate()
vertices = polydata.points
triangles = polydata.faces.reshape(-1, 4)[:, 1:]
print(f"batch with {len(triangles)} triangles")
kwargs = {"triangles": triangles, "vertices": vertices}
for name, val in inputs.items():
t = time_it(val["func"], **{k:v for k,v in kwargs.items() if k in val["names"]})
res = {"name": name, "time": t, "triangles": len(triangles)}
print(res)
results.append(res)
df = pd.DataFrame(results)
df["num of triangles"] = df["triangles"].astype(str) + " "
df["time [ms]"] = df["time"]*1000
fig = px.bar(
df,
x="num of triangles",
y="time [ms]",
# facet_col="variable",
color="name",
#log_x=True,
log_y=True,
facet_col_spacing=0.05,
barmode="stack", # ['stack', 'group', 'overlay', 'relative']
)
fig.show() |
|
to test some memory issue: import magpylib as magpy
import matplotlib.pyplot as plt
import numpy as np
import pyvista as pv
Nt = 10000 # target number of triangles
r = int(np.sqrt(Nt/2))
sphere = pv.Sphere(theta_resolution=r, phi_resolution=r)
magnet = magpy.magnet.TriangularMesh.from_pyvista(
magnetization=[0, 100, 0],
polydata = sphere,
)
N = 30
xs = np.linspace(-1e3, 1e3, N)
ys = np.linspace(0, 1e3, N)
grid = np.array([[(x, y, 0) for x in xs] for y in ys])
%%time
Np = np.prod(grid.shape[:-1]) # number of points
Ns = 100 # number of subsets
slices = [slice(i * Np // Ns, (i + 1) * Np // Ns) for i in range(Ns)]
B = np.concatenate(
[magpy.getB(magnet, grid.reshape(-1,3)[sl]) for sl in slices]
).reshape(grid.shape)
%%time
B1 = magpy.getB(magnet, grid)
np.testing.assert_allclose(B, B1) |
|
this is completed with #598 |

















Discussion how the FacetBody class should look like.
TBD
from previous posting:
The FacetBody class
This class should be quite similar to the TriangularMesh class that @Alexboiboi introduced in the original facet branch, but it is still work in progress and features should still be discussed. The idea is that triangular meshes can be handed over easily, that inside-outside checks will be performed by default ( but can be turned off), and that facet orientation is automatically fixed by default (but can also be turned off).
Parameters: magnetization (the usual) and vertices shape (n,3) and triangles shape (n,3)
It seems to be the common choice in defining meshes through a list of unique vertices, and a second list of indices that define the cells. This is done in pyvista, scipy, pymesh, gmesh, ... . Since we already used vertices in other classes I would use this again for the points, and I really like triangles for the index list.
The text was updated successfully, but these errors were encountered: